
- Pymol tutorial books how to#
- Pymol tutorial books install#
The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC. You can select other formats such as MOL2, Maestro, XYZ, and so on. Go to File -> Export Molecule -> PDB Options -> Write CONECT records for all bonds -> Save. After finishing the structure, you can save it as a PDB file. Click on the amino acids you wish to add to the structure. Repeat the above steps and then switch to the ‘Protein’ tab on the Builder window. Similarly, you can add some amino acids to the structure. You can undo any step by pressing Ctrl+Z, create bonds, clear up the whole screen, add charge, set up a cycle bond (for that click on Cyclewritten in front of ‘ Bonds:’ in the builder window) and so on. Now add other atoms or bonds as per your requirement. Pymol tutorial books how to#
Learn how to use autodock software which is used for molecular docking this a tutorial for basic understanding how to run a docking. For autodock 1 and/or autodock vina 2) are autodock tools (adt). For visualization of molecular structures, amdock uses pymol. Now click on the blank space on the Pymol screen. Morris and stefano forli the scripps research institute.Everyday low prices and free delivery on eligible orders. Contents 1 The PyMOL Interface 1.1 About the command line 2 Getting started: explore a protein 2. This guide is intended to introduce the PyMOL interface and basic tasks without leaving the mouse behind. Get Free Protein Ligand Interactions Volume 53 Store. Although PyMOL has a powerful and flexible interface, it is complex, and can appear daunting to new users.
 Click on the first tab, i.e., Create As a New Object. PyMOL Tutorial: Modeling the SARS-CoV-2 RBD Interactions with ACE (COVID-19 Coronavirus Proteins) Molecular Docking: AUTODOCK VINA Tutorial- PART 1 Topic 6.2 - Ligand. You will see three tabs: (i) Create As a New Object, (ii) Combine w/ Existing Object, and (iii) Done. Click on a structure, for instance, benzene. Choose whatever chemical structure you want, for example, benzene, carbon, oxygen, and so on. After clicking on it, you will see a small window consisting of some chemical structures, atoms, and bonds. In the last row, you will see an option namely, Builder. Click on it. Go to the top-right corner where you can see some options such as Reset, Zoom, Draw, etc. Open Pymol on Ubuntu either by typing pymol on the terminal and on Windows by double-clicking the app.
Click on the first tab, i.e., Create As a New Object. PyMOL Tutorial: Modeling the SARS-CoV-2 RBD Interactions with ACE (COVID-19 Coronavirus Proteins) Molecular Docking: AUTODOCK VINA Tutorial- PART 1 Topic 6.2 - Ligand. You will see three tabs: (i) Create As a New Object, (ii) Combine w/ Existing Object, and (iii) Done. Click on a structure, for instance, benzene. Choose whatever chemical structure you want, for example, benzene, carbon, oxygen, and so on. After clicking on it, you will see a small window consisting of some chemical structures, atoms, and bonds. In the last row, you will see an option namely, Builder. Click on it. Go to the top-right corner where you can see some options such as Reset, Zoom, Draw, etc. Open Pymol on Ubuntu either by typing pymol on the terminal and on Windows by double-clicking the app. Pymol tutorial books install#
If not, then read this article to install Pymol on Ubuntu.
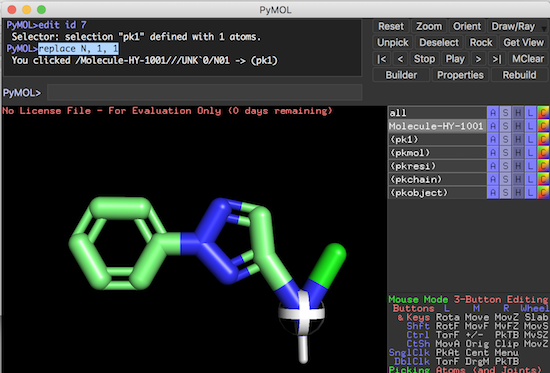
In this article, we will explain how to draw molecules in Pymol.Īssuming you have Pymol installed on your system.
Pymol has many functions that you keep finding on exploration.







 0 kommentar(er)
0 kommentar(er)
